We’re happy to share our latest publication in Cell:
“BRAIN-MAGNET: A functional genomics atlas for interpretation of non-coding variants”,
a collaboration between BIGR and the Clinical Genetics department.
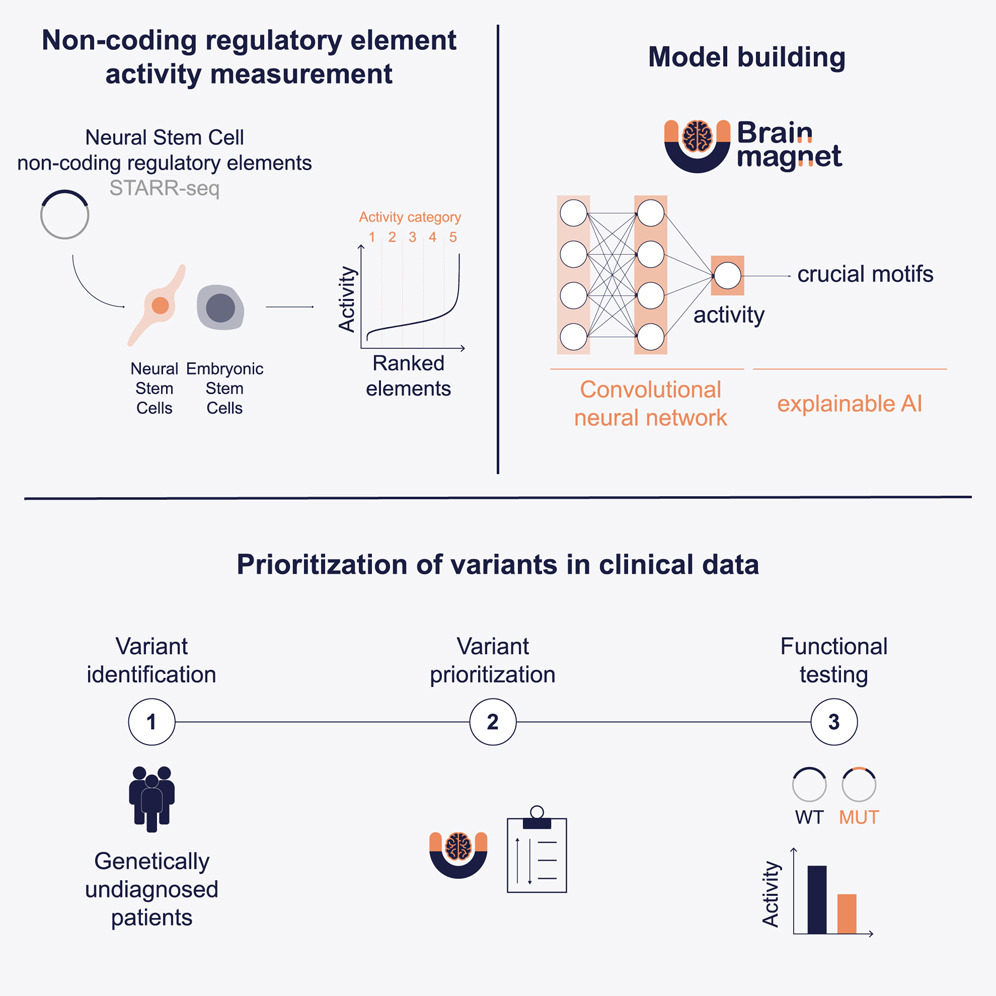
In this work, they (first author Ruizhi Deng (Clinical Genetics department) with co-authors, in particular, Gennady from the BIGR) introduce BRAIN-MAGNET, a convolutional neural network trained on more than 148,000 functionally tested non-coding regulatory elements. Leveraging this large functional genomics atlas, BRAIN-MAGNET can predict enhancer activity directly from DNA sequence and pinpoint nucleotides that are critical for regulatory function in the developing human brain.
By combining high-throughput functional assays with deep learning, BRAIN-MAGNET helps to bridge the gap between non-coding genetic variation and disease. The model supports:
- Prediction of enhancer activity from primary DNA sequence in brain-relevant cellular models;
- Fine-mapping of GWAS loci for common neurological traits;
- Prioritization of rare non-coding variants in patients with unresolved neurogenetic disorders.
Within our group, Gennady is the co-author affiliated with BIGR, contributing expertise on AI and statistical genomics at the interface of radiology, epidemiology, and functional genomics.
The paper is now available online in Cell (Online ahead of print, November 19, 2025; DOI: 10.1016/j.cell.2025.10.029).