About the team
We are a team of Research Software Engineers and Researchers committing ourselves to designing and developing an infrastructure and services for enabling state-of-the-art research using Medical Images. The infrastructure and services we develop also have the aim to make the translation of AI methods to clinical use more straightforward.
We have created a reference architecture that can support the data management for imaging cohort or population studies, by automating large parts of the data handling and data analysis from scanner to extraction of Quantitative Imaging Biomarkers. It is, however, not limited to this, the components in the infrastructure can for example be used to create curation and annotation worflows, secure data export from clinical centers to central storage services, automated analysis workflows, to name a few. In the figure below you can appriciate a schematic overview of the components of our infrastructure and how they can be connected.
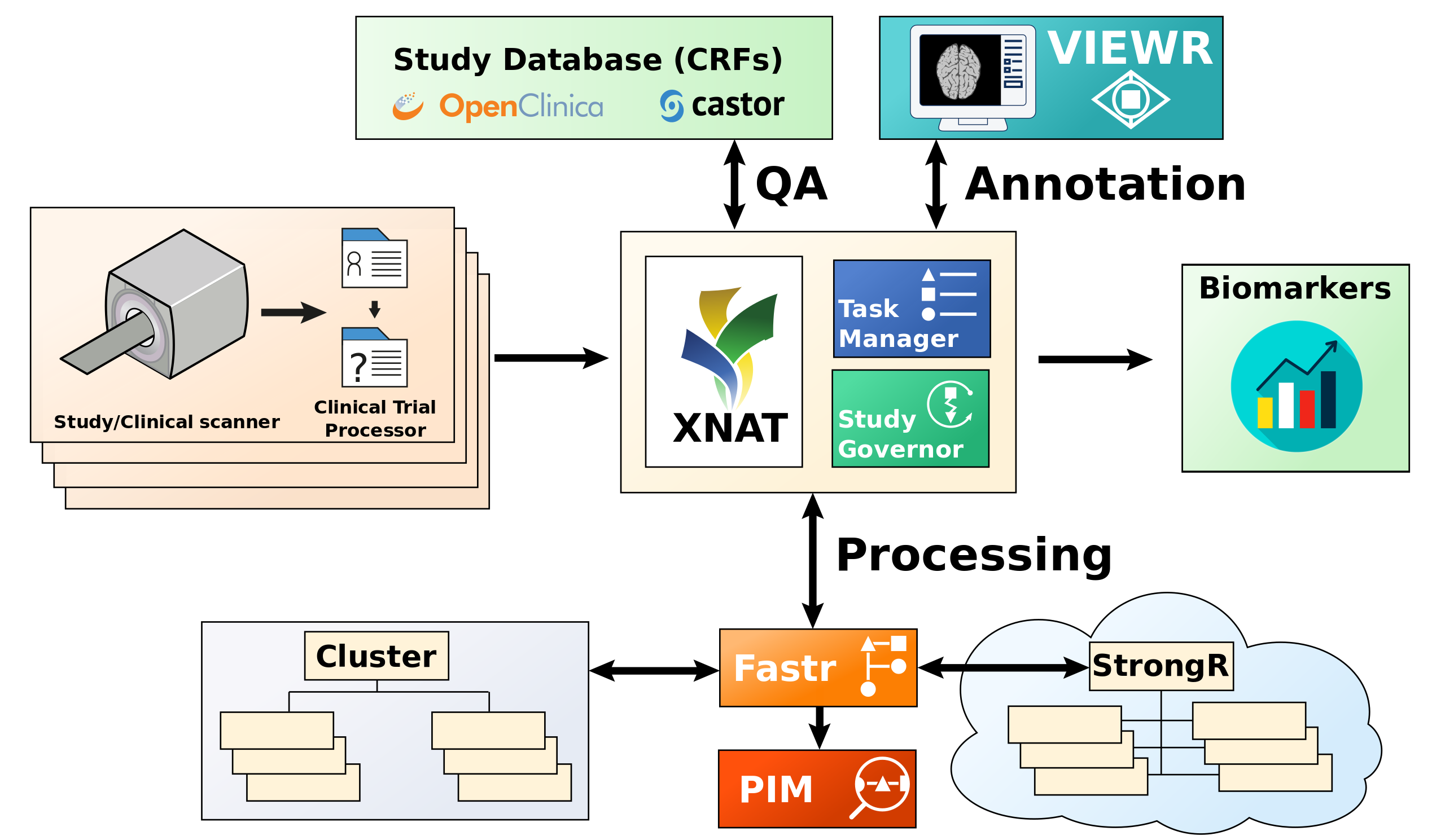
Overview of the image analysis infrastructure. On the left side are the centers where the data is acquired and anonymized. In the center is the XNAT archive, the studygovernor and a tasksupervisor. On the top are the study database and the viewing/annotation applications for manual annotations. On the bottom is fastr and a compute cluster/cloud for automated analysis. Finally all derived data is collected and exported in a format the study desires. The different components interface with each other through REST APIs.
Desing Principles
While developing the components and services that are in our infrastructure we keep to the following principles:
- Improves robustness
- Increases reproducibility
- Increases data consistency
- Is automated where possible
- Consists of modular building blocks
Anonymization and data transport
Generally we use the Clinical Trial Processor (CTP) for anonymization, secure data transport and allocating data to a project. CTP allows the creation of processing pipelines for DICOM scans, in which scans are received, processed and forwarded to another DICOM receiver.
Medical imaging data storage using XNAT
At the heart of the infrastructure is the storage of the medical images. For this we use XNAT, developed by Flywheel. We host our own servers within our institute, but also maintain the Dutch & European XNAT server operated by Health-RI and Euro-BioImaging. We also created and maintain a Python library to communicate with XNAT called xnatpy. XNAT is used to standardize the image storage, making it easier to automate. XNAT also allows for Poject based data access management.
Compute infrastructure
In medical imaging the extraction of a biomarker from an image is mostly not a single step, but an entire processing pipeline. To formalize these pipelines and help executing them in a consistent manner on compute clusters or the cloud, we created the fastr workflow software. To track the progress and status of complex pipelines our colleagues in the Leiden University Medical Center (LUMC) created the Pipeline Inspection and Monitoring (PIM) platform.
Data and workflow management services
The data and workflow management services are a collection of tools and services to manage the data flow, including manual interaction with the data, in an automated fashion. The key components are the Study Governor for automatically managing the data flow and the Task Supervisor for task based manual interaction with the data.
Manual inspection and annotation tasks
The ViewR is a tool to interact with the data on XNAT by researchers based on the tasks served by the tasksupervisor. The layout and forms in the ViewR are determined by templates supplied by the tasksupervisor. This way the ViewR adapts itself for the task to be performed.






