About the research topic
EOSC4Cancer will make diverse types of cancer data accessible: genomics, imaging, medical, clinical, environmental and socio-economic. It will use and enhance federated and interoperable systems for securely identifying, sharing, processing and reusing FAIR data across borders and offer them via community-driven analysis environments.
EOSC4Cancer’s well curated data sets will be essential input for reproducible and robust analytics and computational methods – including machine learning and artificial intelligence. EOSC4Cancer’s five use cases will cover the patient journey from cancer prevention over diagnosis to treatment, laying the foundation of data trajectories and workflows for future European Cancer Mission projects.
Work package 2: Data Harmonization
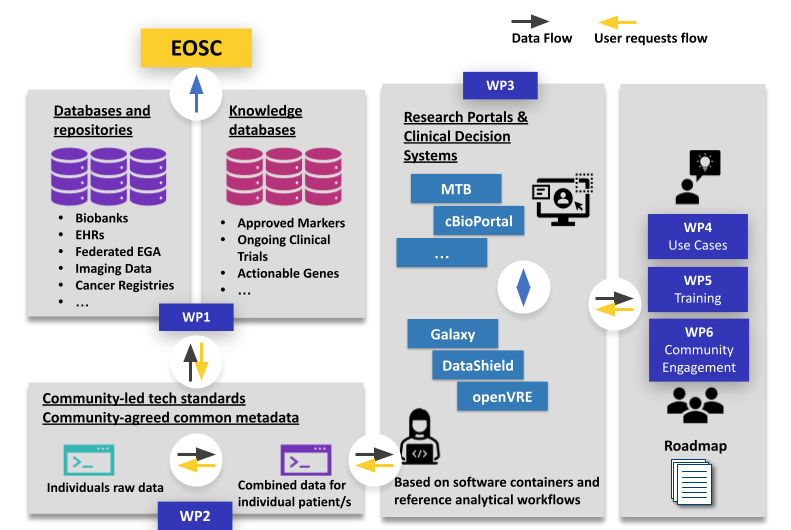
Image from https://zenodo.org/records/10067318.
We lead work package 2, which develops data harmonisation efforts to make cancer research more reproducible across all Member States. It will simplify complex data queries and provide empirical evidence of the interoperability methodology for future studies. Specifically, it will create a model for clinical information retrieval across common nodes; provide guidelines on models, metadata, terminology and provenance; demonstrate proof-of-concept of harmonised data models, terminologies and tools, as well as develop methods to support data aggregation.
Like this, the workpackage will provide mechanisms to facilitate:
- Clinical metadata mining, archiving and interoperability
- Cancer multi-omics and imaging data interoperability
- Federated discoverability for assembling virtual cancer cohorts
An important contribution from BIGR is the integration of XNAT and cBioPortal to enable integrated analysis.


